Context
Breast, ovarian, endometrial and lung tumor samples are routinely homogenized and processed for DNA in cancer research. In addition to local diagnostic requirements, DNA obtained from tumour samples is submitted to the 100,000 Genome Project that aims to use Whole Genome Sequencing (WGS) technique on patients, plus their families, with a rare disease or cancer. This project imposes high standards of DNA quantity and fragment length quality.
In this study, the Minilys tissue homogenizer was evaluated for tumor tissue sample homogenization and results were compared to those obtained following a manual sample homogenization method. The DNA yield and quality as well as hands-on time required were compared between the two methods.
Materials
Samples: 4mm punch biopsies of frozen specimens
Buffer: Proteinase K buffer
For Minilys Method: Minilys homogenizer and 2mL CK28-R Precellys lysing kit.
For manual method: Mini plastic disposable pestle and mortar (optional, a razor blade or scalpel)
Protocol
Manual method: The frozen biopsies were manually treated using the mini plastic disposable pestle/mortar. Samples not homogenized satisfactorily, were chopped up using razor blades/scalpels (treatment time: 5 to 10 minutes per sample). Each sample was then split in 2 tubes: one for storage and one for analysis. Tube for analysis was lysed overnight with 20 μl of Proteinase K (at 37°C) followed by a fluid extraction performed on the next day with a standard kit extraction.
Minilys method: The biopsies were placed into Precellys 2ml CK28-R tubes containing 180μl of ATL buffer. The samples were homogenized with Minilys for 2×20 seconds at 5,000 rpm and at the end of the run, 20μl of Proteinase K were then added directly into the tube for lysis (1h at 37°C). After lysis, each sample was split into 2 tubes: one for storage and one for analysis. Fluid extraction was then performed on the tube for analysis with a standard kit extraction.
Results
• The processing time was significantly reduced when using Minilys for homogenization, as well as the post treatment time with Proteinase K (reduced to one hour vs overnight for the manual method).
• The yield of DNA recovery with Minilys was higher in 81% of the samples compared to the manual method. Nine out of eleven samples homogenized by the manual method didn’t exceed the concentration of 15ng/μl while the lowest concentration found in samples homogenized by the Minilys was 3 times higher (49ng/μl). The average DNA yield recovery with the Minilys was 185.7ng/μl compared to 26.8ng/μl for the manual method. Therefore, only 1 sample needed to be treated.
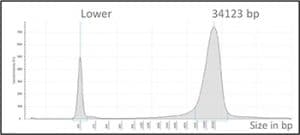
• All DNA samples obtained with the Minilys showed good quality, including excellent fragment length (Figure 1) meeting the 100.000 genome requirements of >60% of fragments with a minimal length of 23kbp.
Figure 1. Fragment length characterization using the TapeStation instrument (Agilent)