Context
Dadih is a traditional Indonesian fermented buffalo milk drink with numerous health benefits associated with several probiotic strains. The current process of dadih production is still mostly artisanal and does not rely on good hygiene practices. The Dadih Initiative was started in Indonesia in 2017 to standardize and optimize production and quality control of dadih. However, before expanding the production of dadih, additional insights into the nature of microorganisms present in the final product are needed. With the advent of next generation sequencing technologies (NGS), it is now possible to explore the microbiota composition of dadih samples with an unprecedented level of complexity. In this work, the microbiota composition of artisanal dadih samples was determined using 16S‐rRNA‐gene amplicon‐sequencing of the V3-V4 region. Several sites of collection and production methods were evaluated, including samples made from pasteurized or raw milk, and using back‐slopping (a practice which involves using previous fermentation as a starter culture) or not.
Materials
- Precellys 24 tissue homogenizer (Bertin Technologies, Montigny‐le‐Bretonneux, France)
- Quick‐DNA™ Fecal/Soil Microbe Miniprep Kit (Zymo Research, Irvine, CA)
Protocol & Results
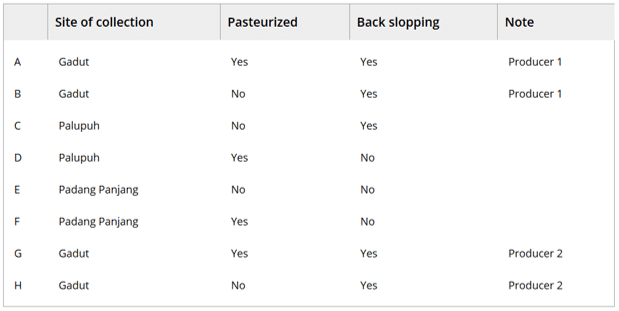
- Dadih samples were collected from two different regions in Bukittinggi city (Gadut – 2 separate producers – and Palupuh), and in Padang Panjang town. There were differences in back‐slopping practice, where previous fermentations were used as a starter culture (five samples) or not (three samples). Table 1 gives an overview of the different samples obtained. Duplicate samples were stored frozen.
- DNA extraction: the samples were centrifuged at 14 500 g and subsequent DNA extraction from the pellet was performed using the Quick‐DNA™ Fecal/Soil Microbe Miniprep Kit (Zymo Research, Irvine, CA) according to manufacturer’s instructions, and the Precellys 24 tissue homogenizer (Bertin Technologies, Montigny‐le‐Bretonneux, France), with 3 cycles of 30s at 6500 rpm, with 5 min cooling on ice in between.
- PCR‐amplifying the V3–V4 region of the 16S rRNA gene and next generation sequencing: Illumina 16S rRNA gene amplicon libraries were generated and sequenced at BaseClear (Leiden, the Netherlands). In short, barcoded amplicons from the V3–V4 region of 16S rRNA genes were generated using a 2‐step PCR.
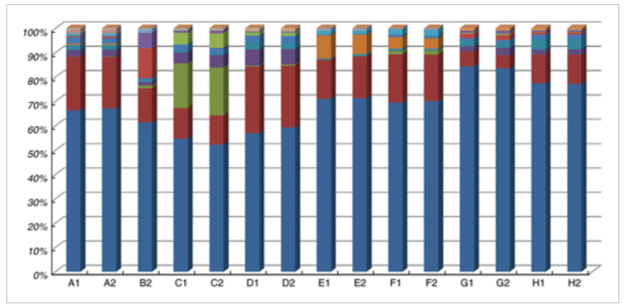
- Sequence processing and analyses: The sequencing run was analysed with the Illumina CASAVA pipeline (v1.8.3) with demultiplexing based on sample‐specific barcodes. Results can be found in Figure 1.
Table 1: Origin and characteristics of the collected dadih samples. From (1)
Figure 1: Relative abundance of major OTUs (at least 0·1% in any of the samples) in the duplicate dadih samples (coded according to Table 1). Legend for microbial OTUs (at genus level, or higher if not available at genus level, indicated by f__: family, o__: order. When the same names are given, these are still different OTUs): Lactococcus ; Klebsiella ; f_Bifidobacteriaceae ; Leuconostoc ; f__Enterobacteriaceae ; Streptococcus ; f__Enterobacteriaceae ; f__Lactobacillaceae ; Lactobacillus ; f__Lactobacillaceae Acinetobacter ; f__ Oxalobacteraceae ; Vagococcus ; Acetobacter ; Proteus ; f__Enterococcaceae ; o__Lactobacillales, and Corynebacterium . From (1)