RNA extraction
RNA isolation can often be challenging, particularly when working with complex samples. Common difficulties included low yield, low purity, RNA degradation, and DNA contamination. Below, Bertin’s team of scientists have shared best practices for homogenization protocols that allow for successful isolation of high-quality RNA.
RNA is a molecule involved in the regulation and expression of genes. Since the discovery of the role of RNA as the key intermediate between DNA and proteins, RNA extraction has become a crucial activity in molecular biology. However, isolating RNA from samples is widely considered to be a tricky endeavor. Due to its single-stranded structure that differs from DNA double strands, RNA is notoriously unstable and sensitive to temperature elevations. RNases, a group of enzymes that degrades RNA molecules, are often found in high quantity in the environment. Common challenges of RNA extraction include RNA degradation, low yield, low purity, and DNA contamination. Furthermore, some sample types have properties that require special treatments. For example, animal tissues contain endogenous RNAase that must be inactivated immediately after harvesting to prevent RNA degradation. Plant and fecal samples can contain inhibitors (e.g. humic acids and polyphenolics) that can co-precipitate with RNA and affect downstream analysis.
As the first step of RNA extraction, sample homogenization plays a crucial role in maximizing the yield and quality of extracted RNA. Complete sample homogenization is essential for the extraction procedure to go smoothly. Indeed, incomplete homogenization followed by column-based extraction methods can cause column clogging as well as other issues. At Bertin Technologies, our team of scientists has developed a bead-beating based homogenizer range, the Precellys, to make RNA extraction an easy and painless process.
Below, our scientists share their tips to design optimized homogenization protocols to get high-yield, high-quality RNA.
Tips for a successful RNA extraction
Stabilizing RNA in the sample
As soon as the sample has been collected, RNA should be protected against degradation caused by heat or RNAses. A common method to inactivate RNAse is the use of buffers containing chaotropic agents such as guanidinium isothiocyanate, guanidinium chloride, sodium dodecyl sulphate (SDS), sarcosyl, and urea. TRIzol or RNAlater can be used to maintain RNA integrity. Samples can also be flash-frozen in liquid nitrogen to avoid RNA degradation.
Working in an RNAse-free environment
Working in an RNase-free environment is essential for successful RNA extraction. If possible, it is best to define a specific benchtop area for your RNA extraction, which should be kept as clean as possible. Working areas should be decontaminated with RNase surface decontamination solution before starting any extraction. It is also important to carefully choose the materials used when attempting RNA extraction. RNase-free disposable plasticware is highly recommended. Precellys lysing kits are single-use, and certified DNAse/RNase free. Other non-disposable plastic material can be treated with a 0.1 M NaOH 1 mM EDTA solution and rinsed with RNase-free water.
We highly recommend wearing sterile disposable gloves and changing them after any contact with material not part of your RNase-free working zone. We also recommend wearing a mask and protective goggles while working. Finally, it is important to make sure that all reagents used in your workflow are RNase-free and use DEPC-treated water.
Temperature control
Heat can cause RNA degradation. Bead-beating homogenization can generate mechanical heat, so the temperature of the sample during the homogenization must be controlled as much as possible. To address this challenge, Bertin Technologies have developed the Cryolys Evolution module, an automated cooling system that enables researchers to control the temperature of their sample during the whole homogenization process. For most RNA extraction workflows, our scientists recommend using the Precellys Evolution homogenizer with Cryolys Evolution set at 4°C during the homogenization step.
Choice of Precellys lysing kit
The appropriate lysing matrix should be chosen to ensure complete sample homogenization and maximizing the recovery of high-quality RNA. Please consult our list of lysing kits with recommendations for various kind of samples.
If you have more questions, do not hesitate to contact our field applications specialists, they will be happy to recommend a suitable lysing kit and protocol.
Ratio tissue sample/RNA extraction buffer
Another key factor for a successful RNA extraction is to identify the quantity of sample that your kit can handle. Read carefully the guidelines of your RNA extraction kit and avoid sample overload that will affect RNA yield and quality.
Example:
Guanidinium thiocyanate solutions: 100 mg tissue sample, 5-10 x 106 cells (animal/plant) or 1×107 bacteria for 1mL of buffer.
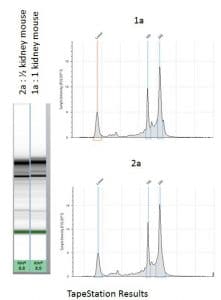
Figure 1 : RNA extracted from whole mouse kidneys. Precellys Evolution and Cryolys Evolution program: 6500 rpm, 20s x 2, pause: 10s at 4°C, with lysing kit CK14 (1.4mm ceramic beads). Following homogenization, the homogenate was transferred to a new 2ml tube and beads were washed with 200ul of Trizol. The resulting 1.2 ml were submitted to a standard Trizol extraction. RNA integrity was checked respectively with TapeStation (Agilent). The resulting high-quality RNA is suitable for RNA sequencing.
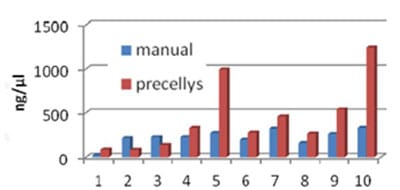
Figure 2 : Extracted RNA yield from subcutaneous human adipose tissue obtained with Ultraturax T8-IKA (manual) and Precellys methods. Precellys 24 dual program: 6000 rpm, 2x15sec, 20s break on ice, lysing matrix: CKMix (mix of 1.4mm and 2.8mm ceramic beads). Buffer: RLT+ß-mercaptoethanol. 10 samples were analyzed, with Nanodrop ND-100. An increase in the amount of extracted RNA is observed with Precellys .
From: INSERM UMRS U872 (Eq7) Nutriomic, Nutrition andEndocrinology Department, Pitié-Salpêtrière Hospital, Paris, France. (V. Pelloux).